Protein Synthesis and Maturation
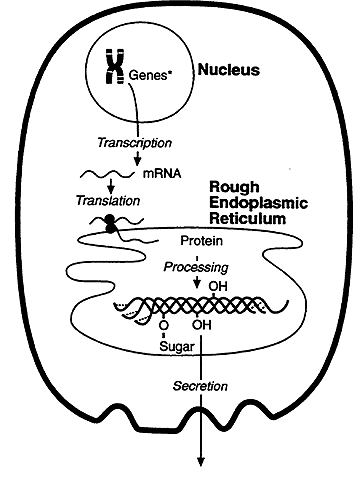
Genetic information flows from DNA to RNA to protein. DNA encodes the information required for synthesis of proteins and a copy of the encoded information is transcribed and processed into messenger RNA (mRNA). The information carried by the mRNA directs the synthesis of proteins, this process is called translation. Translation takes place on the surface of particles called ribosome's.
The Genetic Code
The genetic code is a system of specific base sequences that specify which amino acids are to be used for the synthesis of a protein during translation. The genetic code is comprised of codons which consist of a triplet of bases and specify a specific amino acid. There are 64 codon sequences, sixty-one specify amino acids and three direct the termination of translation. Since there are 20 naturally occurring amino acids more than one codon can specify an amino acid. For example, isoleucine is specified by three codons AUU, AUC and AUA. Codons that specify the same amino acid are called synonyms. The genetic code is called degenerate because synonymous codons generally differ only in the third base. The three codons, UAA, UAG and UGA are stop or nonsense codons and direct the termination of translation.
|
Ala/A |
GCU, GCC, GCA, GCG |
Leu/L |
UUA, UUG, CUU, CUC, CUA, CUG |
|
Arg/R |
CGU, CGC, CGA, CGG, AGA, AGG |
Lys/K |
AAA, AAG |
|
Asn/N |
AAU, AAC |
Met/M |
AUG |
|
Asp/D |
GAU, GAC |
Phe/F |
UUU, UUC |
|
Cys/C |
UGU, UGC |
Pro/P |
CCU, CCC, CCA, CCG |
|
Gln/Q |
CAA, CAG |
Ser/S |
UCU, UCC, UCA, UCG, AGU, AGC |
|
Glu/E |
GAA, GAG |
Thr/T |
ACU, ACC, ACA, ACG |
|
Gly/G |
GGU, GGC, GGA, GGG |
Trp/W |
UGG |
|
His/H |
CAU, CAC |
Tyr/Y |
UAU, UAC |
|
Ile/I |
AUU, AUC, AUA |
Val/V |
GUU, GUC, GUA, GUG |
|
START |
AUG |
STOP |
UAA, UGA, UAG |
Translation is usually initiated with the codon AUG, which specifies Methionine. The remainder of the message is read sequentially, one codon at a time, the codons do not overlap, translation ceases when a stop codon is encountered.
For example, the DNA sequence 5' TACGGTGGATATCCCATC 3' is first transcribed into mRNA 5' AUGCCACCUAUAGGGUAG 3'. This mRNA is then translated into Met-Pro-Pro-Ile-Gly.
Amino Acid Activation
Amino acids must be activated for translation to occur. Activation ensures that the correct amino acid will be recognized and that there is sufficient energy for peptide bond formation. Activation is the covalent coupling of amino acids to specific adapter molecules. The adapter molecules are called transfer RNA (tRNA). There is at least on tRNA for each of the 20 naturally occurring amino acids. The tRNA recognize the codons carried by the mRNA and position them to facilitate peptide bond formation.
Step 1
Amino acid + ATP ----------> Amino-AMP-enz + PPi ; enzyme: Aminoacyl tRNA Synthase
The amino is linked via the 5' position to the ribose on the ATP, liberating PPi. Notice the amino-AMP complex remains bound to the enzyme, aminoacyl tRNA synthase.
Step 2
Amino-AMP-enz + tRNA ----------> amino-tRNA + AMP + Enzyme
The amino group is enzymatically transferred to the 3' terminal adenosine of tRNA, liberating the enzyme and AMP.
Over-all Equation:
Amino acid + ATP + tRNA ----------> Amino-tRNA + AMP + PPi
Notice two high energy phosphate bonds are used to form Amino-tRNA. A tRNA molecule that is linked to an amino acid is said to be charged.
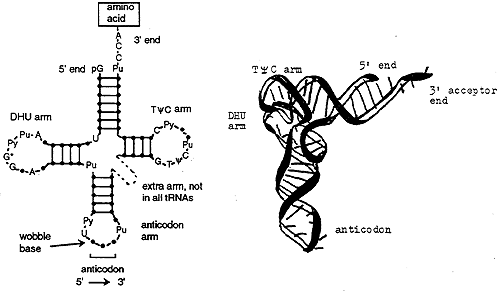
tRNA Structure and Function

All tRNA's are similar in structure. The TyC arm participates in binding of the charged tRNA to a site on the ribosome where protein synthesis occurs. The DHU (or D) arm is necessary for recognition by the proper aminoacyl tRNA synthase (the enzyme). The acceptor end is at the 3' terminus and ends in the sequence CCA. The anticodon arm consists of seven nucleotides, the sequence of which is read 3' to 5' (opposite convention to the usual 5' to 3').
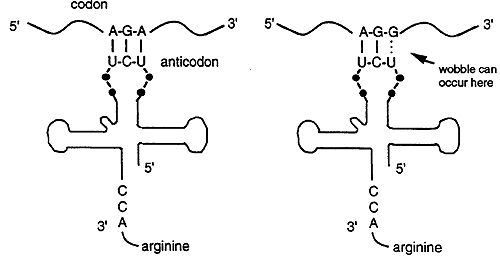
Each anticodon of a tRNA can base pair with a complementary codon on the mRNA. For example, Arginine is specified by two codons, AGA and AGG but there is only one tRNA anticodon for Arg, 3' UCU 5'. The tRNA recognizes and base pairs with either of the two Arg codons. Base pairing occurs between the first two bases of the codon and the anticodon, the third base of the codon does not match. Thus base pairing is not strict for the last nucleotide of the codon anticodon pair, this phenomenon is called wobble.
Ribosome Structure
Ribosome's are found in the cytoplasm, on the outer face of the rough ER and in the mitochondrial matrix. Ribosome's are composed of RNA and proteins. The sedimentation coefficient of eukaryotic ribosomes is 80S. The sedimentation coefficient, S, is a unit of measure that describes how fast a macromolecule will sediment when spun in a high speed centrifuge. Larger molecules generally have larger S values. Eukaryotic ribosomes consist of two subunits: a 60S large subunit and a 40S small subunit. The 60S subunit is comprised of about 45 proteins and three rRNA's that have S coefficients of 5, 5.8 and 28. The 40S subunit consists of about 33 proteins and 18S rRNA's.
Mitochondrial ribosomes are comprised of a large subunit, 16S rRNA and a small subunit 12S rRNA.
Protein Synthesis
Translation of mRNA into a protein requires ribosomes, mRNA, tRNA, exogenous protein factors and energy in the form of ATP and GTP. Translation occurs in three major steps: initiation, elongation and termination.
Initiation
Four major steps are required to initiate translation: ribosome dissociation, formation of a preinitiation complex, formation of the 40S initiation complex and formation of the 80S initiation complex.
Elongation
During elongation the protein is synthesized one amino acid at a time on the 80S ribosome. This process occurs in three major steps: binding of charged tRNA, peptide bond formation, translocation of the growing peptide chain.
Termination
When a stop codon appears at the translation is terminated. There are no tRNA's that recognize stop codons. Instead proteins called releasing factors, eRF, recognize the stop codon. The releasing factors along with peptidyl transferases and GTP catalyze the hydrolysis of the bond between the polypeptide chain and the tRNA. The protein and tRNA disassociate from the site and the ribosome dissociates into the 40S and 60S subunits releasing the mRNA.
Energy Cost for Protein Synthesis
Charging of tRNA: 2 ATP's
Binding of tRNA to Ribosome: 1 GTP
Translocation: 1 GTP
Total Cost: 4 High Energy Phosphate Bonds for each Peptide Bond Formed
Protein Maturation
The rate of protein synthesis is about 6 peptide bonds per minute, thus it takes about 1 to 2 minutes to synthesize an average sized protein. Because mRNA is often several thousand nucleotides in length, the same mRNA molecules can be simultaneously bound by many ribosomes. An mRNA that is bound by multiple ribosomes is called a polysome. Polysomes provide a mechanism for many copies of a protein to be translated from a single mRNA. Polysomes in the cytosol synthesize most of the proteins and enzymes required by the body for intracellular processes such as metabolism.
When protein synthesis terminates, the initiator amino acid, Methionine, will have a free amino group. This end of the protein is the N terminus and the last amino acid of the chain has a free carboxy or C terminus. Protein synthesis thus initiates with the amino terminus and proceeds towards the C terminus. Proteins synthesized on the rough ER are transported across a membrane and into the cisternal spaces between the sheets of the ER where they are packaged for export. To be transported across the membrane the protein is synthesized with a signal or leader sequence on its amino terminus.
After it is synthesized disulfide bonds are formed and the protein folds into its three dimensional state. Some proteins require post-translational modification before becoming fully active. These modifications can include removal of segments using peptidases, addition of phosphate, sugar or lipids to specific amino acids and glycosylation.
Translational Inhibitors
Antibiotics
Streptomycin: prevents tRNA from binding, thus blocking the initiation of translation.
Erthyromycin: binds to the 50S subunit of the prokaryotic ribosome, blocking translocation.
Tetracycline: binds to the 30S subunit of the prokaryotic ribosome and inhibits binding of charged tRNA.
Toxins
Diptheria: catalyzes ADP ribosylation of a His in eEF inhibiting translocation. A few micrograms can kill a human being.
Ricin: cleaves a single adenine from 28S rRNA and inactivates the 60S ribosomal subunit. One molecule can kill an entire cell.
© Dr. Noel Sturm 2014