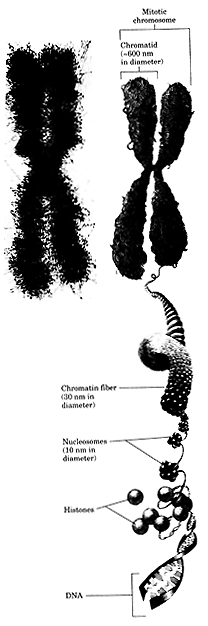
Genome Organization
The human haploid genome consists of about 3 x 109 base pairs of DNA. Genomic DNA exists as single linear pieces of DNA that are associated with a protein called a nucleoprotein complex. The DNA-protein complex is the basis for the formation of chromosomes, virtually all of the genomic DNA is distributed among the 23 chromosomes that reside in the cellular nucleus. A very small fraction of the genome is also found in a 16,000 base pair circular piece of DNA that is found in the mitochondria. The double helical DNA of the chromatin is replicated with the chromatin fiber condensing into discrete bodies, the chromosomes, each consisting of two identical chromatids. The two sister chromatids separate, one moving to each pole of the cell, where they become part of the newly formed nucleus of each daughter cell. The cells that make up most of the body of a multicellular organism, the somatic cells, have two copies of each chromosome and are said to be diploid (2n). Egg and sperm for example, produced by meiosis and having only one copy of each chromosome, are haptoid (n). The DNA of chromatin and chromosomes is bound tightly to a family of positively charged proteins, the histones, which associate strongly with the many negatively charged phosphate groups in DNA. The histones and DNA associate in complexes called nucleosomes in which the DNA strand winds around a core of histone molecules.

Functional Elements and Distribution of DNA within the Genome
The major function of genomic DNA is to carry and store genetic information that is expressed as RNA and then as functional proteins. For gene expression to correctly occur there must be regulatory elements present on the genome and the genome must be faithfully replicated and segregated between daughter cells.
DNA Elements Required for Replication and Segregation of the Genome
Based on studies with unicellular eukaryotes (yeast) at least three types of DNA elements are required for replication and stable inheritance of chromosomes: autonomously replicating sequences (ARS), cetromeres and telomeres. Autonomously Replicating Sequences (ARS) are the sites at which DNA replication is initiated on the chromosomes. Centromeres are DNA sequences that are required for segregation of replicated chromosomes to daughter cells. Telomeres (see "DNA Synthesis" lecture) Telomerase recognizes the tips of chromosomes also know as telomeres. The DNA sequences of telomeres have been determined in several organisms and consist of numerous repeats of a 6 to 8 base long sequence, [TTGGGG]n. Yeast Artificial Chromosomes or YAC's can be constructed by combining large segments of human DNA (50,000 base pairs or longer) with a selectable marker and the three essential elements described above. These artificial chromosomes can then be propagated and amplified in yeast cells. This technology is being used in the sequencing of the human genome.
Unique Sequences
Greater than 50% of the eukaryotic genome consists of DNA that is unique in sequence and the human genome encodes for about 100,000 proteins. The average coding portions of a gene (the exons) consist of about 2,000 base pairs of DNA that is unique in sequence. This number represents less than 7% of the total DNA comprising the human genome and less than 14% of that DNA is unique. Most of the coding sequences are interrupted by from 1 to 50 noncoding sequences or introns. The total length of the introns that interrupt a gene generally far exceeds the total length of the exons. Since sequences that regulate gene expression also account for some of the unique sequences the actual amount of DNA coding for functional gene products is probably less than 3% of the total genomic DNA. The spatial distribution of genes, exons, introns and regulatory sequences along each chromosome is shown below.
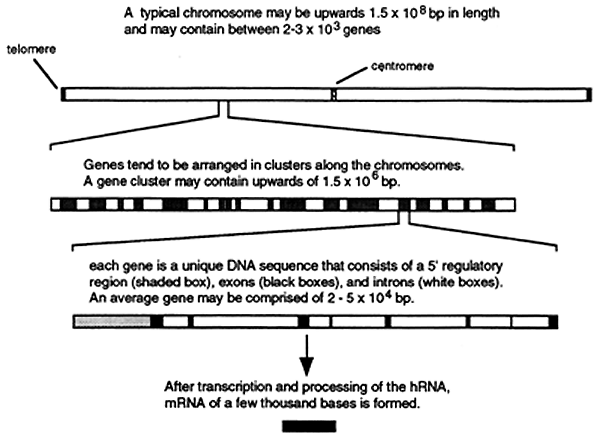
Repetitive Sequences
There are multiple classes of repetitive DNA, two of these classes include: highly repetitive and moderately repetitive DNA. The function of repetitive DNA is not really known but approximately 30% of the human genome consists of repetitive DNA.
Highly Repetitive DNA consists of several different sets of short repeated polynucleotides, generally the repeats range from 5 to 500 base pairs in length and exist in tandem arrays. Highly repetitive DNA comprises about 10-15% of the total genomic DNA, is present in over a million copies and is transcriptionally inactive. Some of the highly repetitive DNA is clustered in structural regions of chromosomes particularly in the cetromeric and telomeric regions.
Moderately Repetitive DNA contains a large variety of repeated sequences ranging from a few hundred to tens of thousands of base pairs with different characteristics. Moderately repetitive DNA can be clustered at specific chromosomal locations or distributed throughout the genome. One type of moderately repetitive human DNA sequence is the rRNA precursor gene. Each rRNA precursor gene is contained in a DNA segment of about 43,000 base pairs. The actual transcript is 13,400 bases which is processed into the mature 28S, 18S and 5.8S rRNA's (see "RNA Synthesis and Processing" lecture). This means that at least 30,000 base pairs are not transcribed and apparently serve as spacer DNA. About 280 copies of the rRNA precursor gene are distributed in clusters on five chromosomes and account for about 0.4% of the genomic DNA.
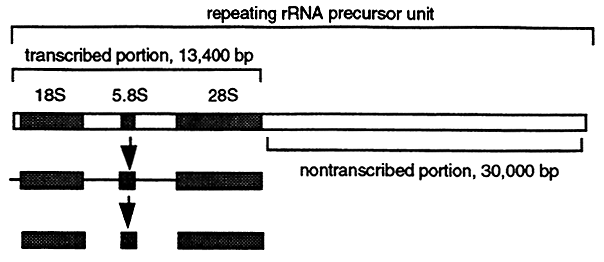
Most types of moderately repetitive DNA are short about 300 base pairs in length, are interspersed with unique sequences, are often transcribed but do not code for gene product.
Chromosomal Structure
Atypical human cellular nucleus is between 5 and 10 mM in diameter and the diploid human genome is over 2 meters long! Obviously to make the DNA fit into the nucleus it must be compacted, think of it as trying to put a piece of thread 6 miles long into a ping-pong ball. Fully compacted DNA can not be transcribed so consequently the cell must be able to selectively expose ARS elements so that replication can be initiated at the correct time in the cell cycle. In order to accomplish all of these tasks, compaction, transcription, replication the DNA is associated with a special set of structural proteins that form a nucleo- or DNA-protein complex called chromatin.
Composition and Structure of Chromatin
Chromatin contains two classes of protein: histones and nonhistone proteins. The overall purpose of histones is to condense the DNA though many nonhistone proteins are involved with transcription, DNA replication and maintenance of chromatin structure.
Histones are the most abundant proteins found in chromatin. There are five major types: H1, H2A, H2B, H3 and H4. The histones are small basic proteins composed mostly of Lys and Arg. The positive charge (basicity) of the histones allows the negatively charged DNA to "wrap" around it forming a nucleosome.
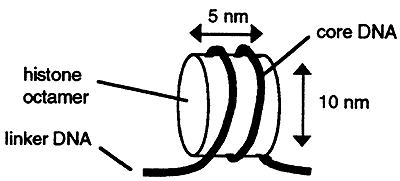
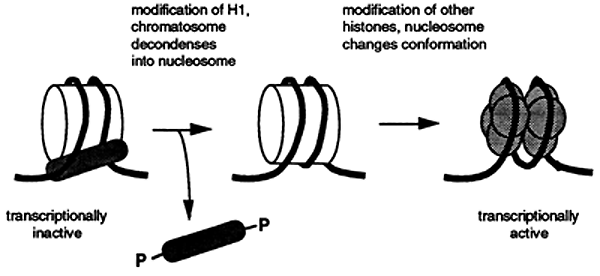
Chromatin consists of a linear chain of nucleosomes each linked to its neighbor by a segment of DNA that is between 20 and 100 base pairs in length. Nucleosomes that are bound to H1 are called chromatosomes. The assembly of nucelosomes is believed to require the participation of the nonhistone proteins, N1 and nucleoplasmin.
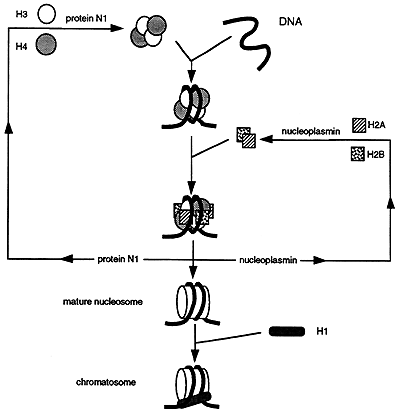
Nucleosome Assembly
The assembly of the nucleosome requires the nonhistone proteins N1, binds to a tetramer of H3 and H4, and nucleoplasmin which binds to dimers of H2A and H2B. The resulting H32H42 tetramer and H2AH2B dimers associate with the DNA while N1 and nucleoplasmin are released and recycled. H1 then adds to the structures forming a chromatosome.
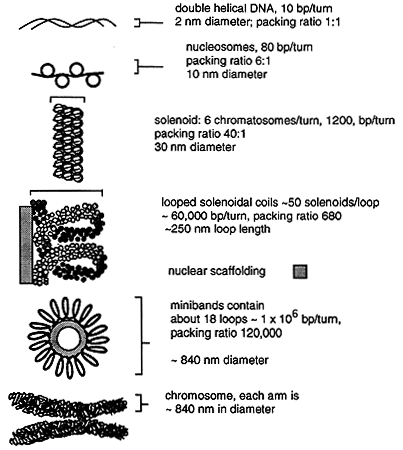
Chromatin can be further compacted into higher order structures including a solenoidal coil with about six chromatosomes per turn and the resulting DNA fibril. The fibril forms loops anchored to a nonhistone protein scaffold, the looped structures forming the interphase chromosomes. During mitosis the looped structure further condense by coiling upon themselves to form minibands. Each miniband is comprised of about 18 loops, each loop containing over a million base pairs. The DNA in these minibands has been compacted by about 10,000 fold! The minibands are arranged along a central axis and form the arms of the mitotic chromosome.
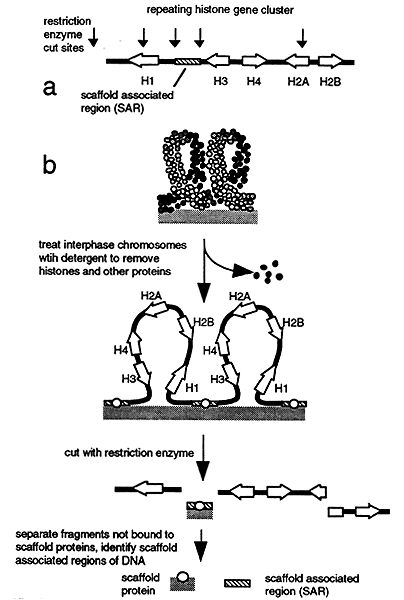
Treatment of mitotic chromosomes with dextran sulfate followed by special detergents strips off the histones and most other proteins. Additional treatment with restriction enzymes cuts most of the DNA which can then be separated from the scaffold. When the scaffolding is then analyzed short segments of DNA are found attached to the scaffolding between genes, not within regions of transcribed DNA. These sequences are called scaffold associated regions or SAR's.
The major scaffold protein is topoisomerase II which regulates the extent of supercoiling in the DNA. Supercoiling, seen in circular DNA (mitochondrial DNA) and nucleosomes (DNA wrapped around something else), results when double stranded DNA twists upon itself. Topoisomerase II maintains the level of supercoiled DNA at a constant value because supercoiling can affect the efficiency of transcription, DNA replication and the integrity of chromatin.
Chromatin Dynamics
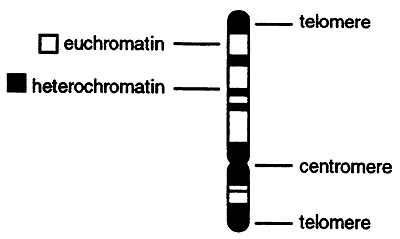
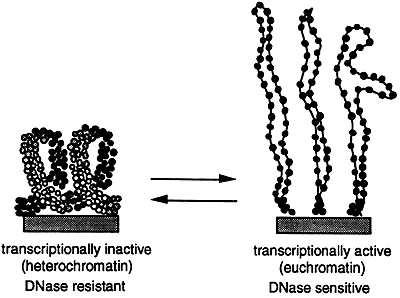
The higher order structure of chromatin varies and is determined by factors such as tissue type, sex and the developmental state of the cell. If chromosomes are stained with a dye and then analyzed microscopically numerous dark bands are seen. The dark bands correspond to the highly condensed and transcriptionally inactive heterochromatin. Heterochromatin is generally found at or near the centromere and telomeres and consists of highly repetitive DNA. The lighter bands are the less condensed, transcriptionally active euchromatin.
In order for DNA replication to occur the chromatin must be dynamically restructured or "decondensed" allowing the replication "machinery" to gain access to the DNA. Transcriptionally active genes are sensitive to digestion by DNase while inactive genes are insensitive to digestion. This suggests that the chromatin has "decondensed" during transcription which also allows access to the DNase.
Numerous subtypes of histones have been identified. Analysis of these histones indicates that histones are subject to chemical modification via: acylation, phosphorylation, ADP-ribosylation and ubiquination. Some of these modified histones appear to be associated with actively transcribing genes suggesting that the modifications may affect the structure of the nucleosome making the DNA more accessible to the enzymes required for regulating and carrying out transcription, replication and repair.
The mitochondrial genome is 16,569 base pairs in length, circular and does not associate with histones. There is no repetitive DNA in the mitochondria so virtually all of the DNA is used to encode 2 rRNA's, 22 tRNA's and 13 proteins.
© Dr. Noel Sturm 2003